目前,国内外市场三文鱼的销售量稳步增加,挪威、芬兰和美国分别以生产大西洋鲑(Salmo salar)、虹鳟(Oncorhynchus mykiss)和阿拉斯加鲑(Oncorhynchus nerka)为主,中国以生产虹鳟为主。虹鳟是鲑科的一员,在世界范围内具有重要的生态价值[4]。其中,三倍体虹鳟因体内有三套染色体、无法进行减数分裂、不能产生精子和卵子以及不存在性腺发育等特点,具有生长快、肉质好、个体大和能够有效防范物种入侵[5]等优势,而成为黄河上游青海段的主要养殖品种,促进青海省冷水养殖产业的效益稳步提升。目前,有关三倍体虹鳟的研究主要集中在能量代谢[6-7]、发育过程[8-9]以及不同发育阶段基因的表达[10-11]等方面,在鱼种鉴定方面鲜有报道。市售的三倍体虹鳟主要以冰鲜、腌制和烟熏的形式存在,急需一种有效的鉴定方法,以利于保护消费者的合法权益,规范市场监管。
随着现代生物学技术的不断发展,鱼类的物种鉴定研究已逐渐从形态学方法转向形态学和分子生物学相结合的方法[12]。DNA作为生物体最核心的信息遗传载体,其遗传序列直接决定了生物体的本质。以DNA为基础的分子检测技术由于具有较高的灵敏度、特异性、可靠性,己成为鱼类物种鉴定的主要检测技术[13]。分子鉴定技术主要包括随机引物 PCR(Arbitrarily primed PCR,AP-PCR)、随机扩增多态性 DNA(Random amplified polymorphic DNA,RAPD)、扩增性简单序列重复(Inter-simple sequence repeats,ISSR)、限制性内切酶切片段长度多态性(Restriction fragment length polymorphism,RFLP)、扩增限制性内切酶片段长度多态性(Amplified restriction endonuclease fragment length polymorphism,AFLP)、单核苷酸多态性(Single nucleotide polymorphism,SNP)和DNA条形码(DNA-barcode)序列分析[14]等。其中,DNA条形码在物种鉴定中具有独特的优势,即可以根据整条鱼、鱼片、鱼鳍、鱼卵或组织碎片的样本识别鱼类物种,仅需要少量样本即可进行DNA条形码检测。此外,即使是加工处理的样品,也可以应用DNA条形码对其进行物种鉴别[15]。目前,DNA-barcode和Mini-barcode技术已被广泛用于鱼种鉴定,其通过扩增部分COⅠ序列作为条形码,以此对物种进行准确鉴定,但该技术应用于近缘物种、杂交种和变种之间的鉴定尚未被明确。本研究以三倍体虹鳟、二倍体虹鳟及其变种金鳟、大西洋鲑、大麻哈鱼(Oncorhynchus keta)和高白鲑(Coregonus peled)为研究对象,利用COⅠ基因对虹鳟进行DNA条形码研究和系统进化分析,探讨COⅠ基因在分子鉴定中的适用性,为虹鳟的物种鉴定提供技术支撑。
1 材料与方法
1.1 材料
三倍体虹鳟、金鳟、二倍体虹鳟和大西洋鲑样品各3条,分别采自青海省海东市、循化县、化隆县和门源县;冰鲜大麻哈鱼和高白鲑样品各3条,分别购自库尔勒爱果电子商务有限公司和哈尔滨龙德食品有限公司。取以上鱼类的背部肌肉组织,置于-80 ℃保存备用。
1.2 DNA提取及引物设计
使用DNA提取试剂盒(TIANGEN,Code No.DP304)提取上述鲑科鱼类肌肉组织基因组DNA,通过紫外分光光度计(Implen,NanoPhotometer N60)测定各样本的浓度和纯度。以质量合格的DNA为模板,分别扩增6种样本鱼的DNA barcode和Mini barcode,扩增产物用1%琼脂糖凝胶电泳检测。通过NCBI数据库下载10种鲑科鱼类CO Ⅰ基因序列(表1),经Clustal X [16]对比后,选取保守区序列,利用Primer Premier 5软件分别设计两对DNA barcode、Mini barcode引物用于虹鳟鉴定(表2),所有引物委托生工生物工程(上海)股份有限公司合成。
表1 鲑科鱼类COⅠ基因信息
Tab.1
| 物种 Species | 登录号 Accession number | 长度/bp Length |
|---|---|---|
| 帝王鲑 Oncorhynchus tshawytscha | KX958414.1 | 1 204 |
| 金腹大麻哈鱼 Oncorhynchus chrysogaster | JX960908.1 | 1 247 |
| 美洲红点鲑 Salvelinus fontinalis | HQ167687.1 | 1 551 |
| 钝吻鲑 Salmo obtusirostris | JX960948.1 | 1 262 |
| 多瑙哲罗鱼 Hucho hucho | JX960904.1 | 1 247 |
| 细鳞鲑 Brachymystax lenok | JX227988.1 | 1 581 |
| 突唇白鲑 Coregonus lavaretus | JX960890.1 | 1 246 |
| 北鲑 Stenodus leucichthys | KT630716.1 | 1 201 |
| 普伦白鲑 Coregonus pollan | JX960898.1 | 1 262 |
| 茴鱼 Thymallus thymallus | JX960975.1 | 1 262 |
表2 PCR引物信息
Tab.2
| 引物名称 Name of primers | 引物序列 Sequence of primers | 产物长度/bp Length of product | |
|---|---|---|---|
| COⅠ-1(DNA barcode) | COⅠ-F24 | 5’-GGGATGACCAAATCTATAACGTGA-3’ | 681 |
| COⅠ-R24 | 5’-CTCAGACCATTCCTATATACCCGAAG-3’ | ||
| COⅠ-F25 | 5’-GATTAATTCCCCTAATAATCGGAGC-3’ | 678 | |
| COⅠ-R25 | 5’-ATGTAAAGTAAGCACGAGTGTCCA-3’ | ||
| COⅠ-2(Mini barcode) | TEYI-0F | 5’-AACCTCCAGCCATCTCTCAG-3’ | 153 |
| TEYI-0R | 5’-CCGGGTCAAAGAAAGTGGTG-3’ | ||
| TEYI-1F | 5’-CCGCCCTGAGTCTACTGATT-3’ | 230 | |
| TEYI-1R | 5’-TGGAGGAAGGAGTCAGAAGC-3’ | ||
1.3 PCR反应体系、反应条件及产物分析
PCR反应体系为20 μL,包括:Premix Taq 10 μL(Takara,Code No.RR902A),DNA模板1 μL,上下游引物各1 μL(浓度为10 μmol/L),ddH2O补充反应体系至20 μL。CO Ⅰ-1 PCR反应条件:94 ℃预变性1 min;94 ℃变性30 s,50 ℃退火40 s,72 ℃延伸1 min,5个循环;94 ℃变性30 s,54 ℃退火40 s,72 ℃延伸1 min,35个循环;72 ℃延伸10 min,4 ℃结束。CO Ⅰ-2 PCR反应条件:94 ℃预变性3 min;94 ℃变性30 s,60 ℃退火30 s,72 ℃延伸40 s,35个循环;72 ℃延伸10 min,4 ℃结束。PCR产物经1%的琼脂糖凝胶电泳检测,以DNA 2000 Marker(Takara,Code No.3427A)判定PCR产物片段大小与引物产物片段大小的一致性。
1.4 测序及数据分析
表3 17个鲑科物种的COⅠ基因信息
Tab.3
| 种名 Species | 登录号 Accession number | 长度/bp Length | |
|---|---|---|---|
| 帝王鲑 Oncorhynchus tshawytscha | FJ999371.1 | 652 | |
| 银鲑 Oncorhynchus kisutch | MG951604.1 | 652 | |
| 虹鳟 Oncorhynchus mykiss | MN850431.1 | 653 | |
| 大麻哈鱼 Oncorhynchus keta | MN850432.1 | 653 | |
| 大西洋鲑 Salmo salar | MN850430.1 | 651 | |
| 花羔红点鲑 Salvelinus malma | EU522414.1 | 652 | |
| 北极红点鲑 Salvelinus alpinus | KJ128606.1 | 648 | |
| 远东红点鲑 Salvelinus leucomaenis | MF503661.1 | 697 | |
| 多瑙哲罗鱼 Hucho hucho | KJ553640.1 | 652 | |
| 哲罗鱼 Hucho taimen | MG951558.1 | 652 | |
| 细鳞鲑 Brachymystax lenok | JX261990.1 | 633 | |
| 远东哲罗鱼 Parahucho perryi | HQ693237.1 | 655 | |
| 秋白鲑 Coregonus autumnalis | EU202649.1 | 650 | |
| 高白鲑 Coregonus peled | MF632325.1 | 637 | |
| 欧白鲑 Coregonus albula | KX457959.1 | 704 | |
| 茴鱼 Thymallus thymallus | HQ961016.1 | 652 | |
| 黑龙江茴鱼 Thymallus grubii | MG951577.1 | 652 | |
2 结果与分析
2.1 PCR扩增及序列分析
表4 各样本DNA质量检测结果
Tab.4
| 物种 Species | 浓度/(ng/μL) Concentration | OD(260/280) | 260/230 | 260 |
|---|---|---|---|---|
| 三倍体虹鳟Triploid rainbow trout | 164.90 | 2.032 | 1.343 | 3.300 |
| 金鳟Golden trout | 197.65 | 2.141 | 1.633 | 3.964 |
| 二倍体虹鳟 Diploid rainbow trout | 119.85 | 2.001 | 1.097 | 2.366 |
| 大西洋鲑Salmo salar | 170.00 | 1.988 | 1.943 | 3.406 |
| 大麻哈鱼Oncorhynchus keta | 198.40 | 2.084 | 1.859 | 3.957 |
| 高白鲑Coregonus peled | 126.05 | 1.982 | 2.030 | 2.507 |
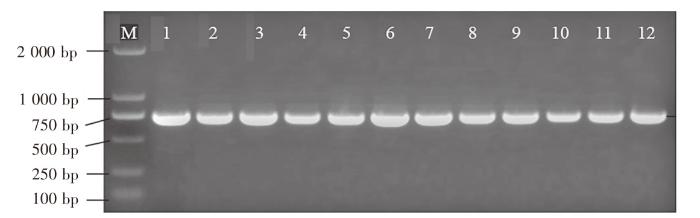
图1
图1
DNA barcode PCR 结果
注:M为DL 2000分子质量标准;1~6使用COⅠ-F24、COⅠ-R24引物;7~12使用COⅠ-F25、COⅠ-R25引物;1、7为三倍体虹鳟;2、8为金鳟;3、9为二倍体虹鳟;4、10为大西洋鲑;5、11为大麻哈鱼;6、12为高白鲑。
Fig.1
DNA barcode PCR results
Notes: M indicated DL 2000 molecular quality standard; 1-6 used COⅠ-F24 and COⅠ-R24 primers; 7-12 used COⅠ-F25 and COⅠ-R25 primers; 1 and 7 were triploid rainbow trout; 2 and 8 were golden trout; 3 and 9 were diploid rainbow trout; 4 and 10 were S.salar; 5 and 11 were O.keta; 6 and 12 were C.peled.
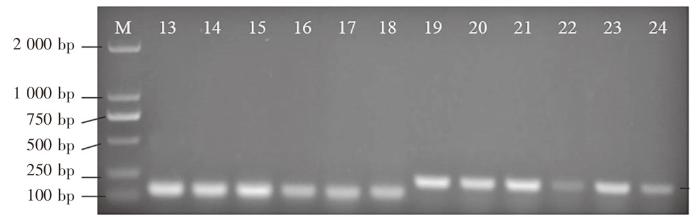
图2
图2
Mini barcode PCR 结果
注:M为DL 2000分子质量标准;13~18使用TEYI-0F、TEYI-0R引物;19~24使用TEYI-1F、TEYI-1R引物;13、19为三倍体虹鳟;14、20为金鳟;15、21为二倍体虹鳟;16、22为大西洋鲑;17、23为大麻哈鱼;18、24为高白鲑。
Fig.2
Mini barcode PCR results
Notes: M indicated DL 2000 molecular quality standard; 13-18 used TEYI-0F and TEYI-0R primers; 19-24 used TEYI-1F and TEYI-1R primers; 13 and 19 were triploid rainbow trout; 14 and 20 were golden trout; 15 and 21 were diploid rainbow trout; 16 and 22 were S.salar; 17 and 23 were O.keta; 18 and 24 were C.peled.
2.2 测序结果分析
将测序得到的DNA barcode和Mini barcode序列拼接处理后,基于NCBI数据库检索比对,结果表明本研究扩增得到的6种鱼的CO Ⅰ基因信息同NCBI数据库中的已知物种信息匹配度与覆盖度均在97%以上。相比于DNA barcode序列,Mini barcode序列的最高匹配度较低,由表5可知,DNA barcode比对结果均为100%,而Mini barcode中只有大西洋鲑比对结果为100%,这可能是因为Mini barcode序列长度较短(153 bp),而长片段(DNA barcode)中个别碱基的缺失、插入及突变会对结果产生较大的影响。通过BLAST发现待测样本高白鲑应为欧白鲑(Coregonus albula),可能存在标签误贴的情况。本研究以CO Ⅰ作为单一靶基因,可对虹鳟和其他鱼种进行区分,但对于近缘物种和变种不能区分,需要寻找新的鉴别手段。
表5 6种鲑科鱼基于DNA条形码技术物种鉴定结果
Tab.5
| 标签名称 Label name | DNA barcode鉴定结果 (序列相似度/%) DNA barcode identification results (sequence similarity) | Mini barcode鉴定结果 (序列相似度/%) Mini barcode identification results (sequence similarity) | 结合两个片段鉴定结果 Combining the identification results of the two fragment |
|---|---|---|---|
| 三倍体虹鳟 Triploid rainbow trout | 虹鳟(100) | 虹鳟(98.69) | 虹鳟 |
| 金鳟Golden trout | 虹鳟(100) | 虹鳟(98.69) | 虹鳟 |
| 二倍体虹鳟 Diploid rainbow trout | 虹鳟(100) | 虹鳟(98.69) | 虹鳟 |
| 大西洋鲑Salmo salar | 大西洋鲑(100) | 大西洋鲑(100) | 大西洋鲑 |
| 大麻哈鱼 Oncorhynchus keta | 大麻哈鱼(100) | 大麻哈鱼(97.06) | 大麻哈鱼 |
| 高白鲑 Coregonus peled | 欧白鲑(100) | 欧白鲑(98.52) | 欧白鲑 |
2.3 系统发育分析
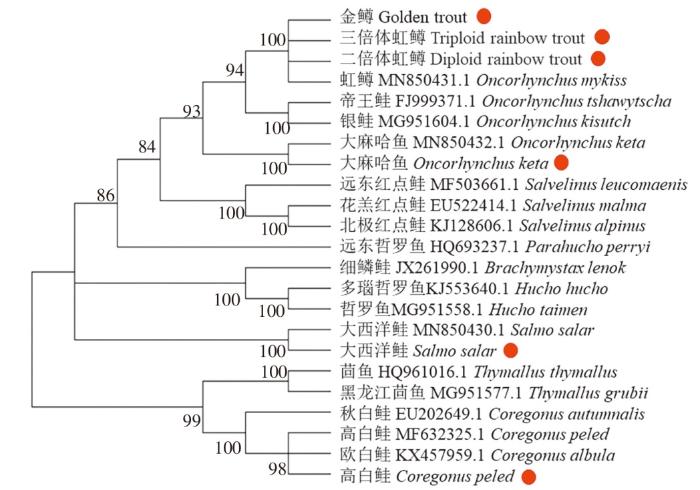
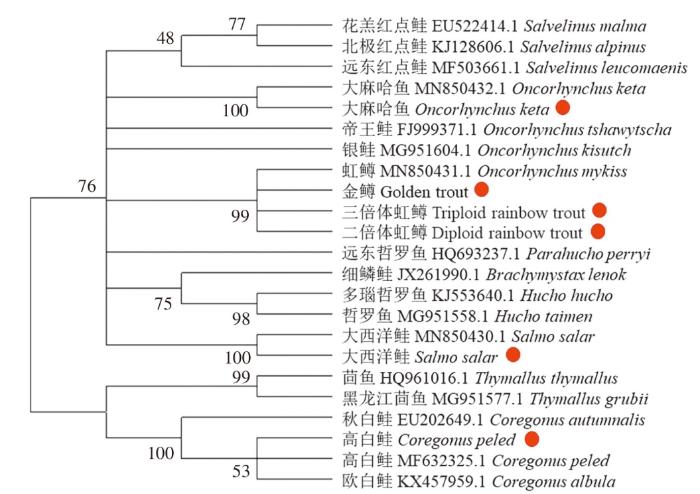
三倍体虹鳟属于鲑科,鲑科有3个亚科11个属,分别基于DNA barcode和Mini barcode构建鲑科鱼类系统发育树(图3、图4)。两个聚类结果基本一致。鲑科17个物种聚为2大支,其中白鲑亚科和茴鱼亚科聚集在一起,鲑亚科聚集在一起,表明白鲑亚科和茴鱼亚科的亲缘关系较近。每个鱼种独立分支,由于三倍体虹鳟是由二倍体虹鳟和四倍体虹鳟杂交出的全雌性虹鳟,金鳟为二倍体虹鳟的变种,因此三倍体虹鳟、金鳟、二倍体虹鳟和虹鳟聚为一支;测序后大西洋鲑和大麻哈鱼的序列也与在NCBI上下载的序列(MN850430.1、MN880432.1)聚集在一起,表明其亲缘关系最近;测序后高白鲑的序列与NCBI下载的欧白鲑、高白鲑聚集在一起;白鲑属(Coregonus)、茴鱼属(Thymallus)聚在一起,麻哈鱼属(Oncorhynchus)、副哲罗鱼属(Parahucho)、哲罗鱼属(Hucho)、红点鲑属(Salvelinus)、鳟属(Salmo)聚在一起,表明此系统发育树可信。
图3
图3
基于DNA barcode构建系统发育树
Fig. 3
Constructing phylogenetic tree based on DNA barcode
图4
图4
基于Mini barcode构建系统发育树
Fig. 4
Constructing phylogenetic tree based on Mini barcode
3 讨论
遗传学方法的优点之一是从任何年龄动物的活体或死体身上收集的一个小组织样本就包含了完整的核和线粒体基因组信息。通过比较基因组选定区域内的中性或保守变异,可以在个体水平上进行鉴定[20]。本研究利用此原理进行采样后虹鳟与其他鱼种的区分。相比较蛋白质检测方法,利用DNA鉴别更为简便,它比蛋白质更耐高温,在不利的环境条件下仍然有可能通过聚合酶链式反应扩增小的DNA片段用于鉴定,DNA还存在于生物体的几乎所有细胞中,可以从任何基质中被提取[21]。DNA分子标记能够直接反映特种DNA分子水平上的差异[22]。DNA条形码技术是利用线粒体基因的短序列来识别物种,应用于特定生物分类群的DNA条形码标记,对物种边界、群落生态学、功能性状进化、营养相互作用和生物多样性保护等研究具有重要意义[23]。DNA条形码技术因准确、经济、高效的特性而备受关注,比较适合鉴别较高经济价值的鱼种[24]。
随着全球贸易鱼类产品的种类和类型的增加,产品标签错误的案例也在增加,而其发生有不同的原因。在某些情况下,当鱼肉具有类似的物理性质(颜色、质地),并且通过加工去除鱼鳍和头部等形态特征时,鱼会被错误地识别为其他种类。Nedunoori A等[25]利用DNA条形码技术结合BOLD识别引擎和本地组装的条形码库,对来自食品店的22个冷冻鱼和鱼片样本进行了误标调查,研究发现白眼鳕鱼被错标为鳕鱼片,罗非鱼无法被准确识别。利用DNA条形码技术,Cline E等[26]对来自美国华盛顿西部商店和餐馆的鲑鱼样本进行了检测,以确定大西洋鲑鱼取代了太平洋鲑鱼,在99个鲑鱼样本中,有11个(11%)是货真价实的;超过38%的餐厅样本被错误标记物种,而有7%的商店样本被错误标记。Clark L F等[27]讨论了鱼类价值链中标签错误和替代问题的范围、公共和私营部门目前使用的条形码以及进一步实施的一些建议。本研究中发现高白鲑存在标签误贴现象,实际应该为欧白鲑,结果同时证明利用DNA条形码技术可以对物种进行鉴定。
在鱼类条形码研究中,通常用于物种鉴定的DNA标记是线粒体DNA(mtDNA)中一个648 bp的区域,被称为COⅠ基因。现有的分子鉴定研究通常使用DNA barcode、Mini barcode或二者与其他基因相结合的方法进行物种鉴定,例如Chang C等[28]、Zahn R J等[29]和Sultana S等[30]分别利用650、130、259 bp的COⅠ基因与141 bp的18S rRNA基因对台湾鱼、鲨鱼和20种商品鱼进行鉴定,结果均显示该方法可行。尽管DNA条形码技术在鱼类物种鉴定方面已逐渐成熟,但其鉴别能力受到许多因素的影响。其中,数据库脆弱性和标本误认是主要因素[31]。本研究中使用681 bp的DNA barcode和153 bp的Mini barcode相结合的方法,对虹鳟及其他鲑科鱼进行鉴定,可以有效地减少在测序过程中因碱基缺失或者错配而产生的误差,可在结果上相互验证,使结果更有说服力;系统发育树的结果表明,此方法可用于虹鳟的鉴定,为其产业的健康发展提供理论依据。
关于鲑科鱼类的系统发育分析,目前常用的方法有NJ法和最大似然法(Maximum likelihood estimate,MLE)[32-33]。本研究使用NJ法对三倍体虹鳟、金鳟、二倍体虹鳟、大麻哈鱼、大西洋鲑、高白鲑和其他17种鲑科鱼类的线粒体基因组构建系统发育树。结果显示,白鲑属、茴鱼属聚在一起,麻哈鱼、副哲罗鱼属、哲罗鱼属、红点鲑属、鳟属聚在一起。本研究所构建的系统发育树与李瑶瑶等[34]根据鲑科鱼cyt b基因构建的系统发育树和孙毅[35]依据鲑科鱼COX1基因构建的系统发育树结果一致,佐证了本研究结果的可靠性。基于序列BLAST和进化树分析发现,检测的6种鱼类样本中高白鲑存在标签误贴的情况,这在鱼类消费市场中常有发生[36-37],故各种鱼类鉴定方法的建立对规范市场具有重要意义。
金鳟为虹鳟的变种[38],二者拉丁名一致,基于线粒体基因组无法被区分,因此需探索其他可对二者进行鉴定的方法。Stephens M R等[39]利用SNP技术对掺入金鳟中的虹鳟进行了区分,本课题组后续将对该方法的适用性做进一步验证。在鱼类中,三倍体是由一个四倍体产生的配子与一个二倍体产生的配子通过受精方式结合的,三倍体化被普遍认为是不育鱼大规模生产最实用、经济和有效的方法[40]。本研究结果显示,三倍体虹鳟、金鳟和二倍体虹鳟的COⅠ基因序列及系统发育树结果一致,且测序发现三倍体虹鳟和二倍体虹鳟线粒体基因组完全一致,故基于mtDNA无法对三者进行区分和鉴定。已有研究发现,依据核基因可鉴定三倍体虹鳟、金鳟和二倍体虹鳟[41-42],但该方法对样本来源要求较高,无法被广泛应用于加工样本的鉴定。综上所述,本研究开发的DNA条形码可将虹鳟与其他鲑科鱼类进行区分,这将为虹鳟物种鉴定及产业的健康发展提供技术支撑。
4 结论
本研究开发的DNA条形码技术可区分虹鳟与其他鲑科物种,但对于三倍体虹鳟、二倍体虹鳟和金鳟之间不能区分,需要寻找新的鉴定方法对变种进行区分。
参考文献
黄河上游地区减贫转向与高质量发展
[J].黄河上游地区是中国贫困易发高发的地区。在2020年打赢脱贫攻坚战背景下,探讨该地区减贫转向特征和高质量发展路径,对实现区域可持续发展具有重要指导意义。本文通过梳理流域地区发展研究和国家战略需求,基于发展地理学视角,构建了面向流域减贫与发展的“五位一体”地理资本体系及其空间整合分析框架。在界定黄河上游地区范围的基础上,阐述了该地区的减贫转向与发展问题。研究发现:黄河上游地区贫困发生率持续下降,贫困人口大幅减少,2020年后将实现减贫转向;地理资本指数呈明显上升趋势,上升幅度高于黄河流域(0.078)和全国平均水平(0.067),但仍受城乡居民收入和城市化水平较低,工业企业研发强度、贸易依存度、专利密度和技术市场水平较弱等方面的制约;该地区与黄河流域和全国比较发展差距在缩小,呈现区域收敛趋势,但欠发达的格局没有改变。在减贫转向和区域收敛的背景下,综合集成构建了该地区可持续减贫及由传统增长向高质量发展转型的路径模式。
The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates
[J].
Threonine modulates immune response, antioxidant status and gene expressions of antioxidant enzymes and antioxidant-immune-cytokine-related signaling molecules in juvenile blunt snout bream (Megalobrama amblycephala)
[J].
Organic and inorganic zinc in the diet of a commercial strain of diploid and triploid rainbow trout (Oncorhynchus mykiss): effects on performance and mineral retention
[J].
Oxidative effect of L-carnitine on energy metabolism in diploid and triploid rainbow trout (Oncorhynchus mykiss): impact on metabolites
[J].
Comprehensive analysis of miRNA-mRNA/lncRNA during gonadal development of triploid female rainbow trout (Oncorhynchus mykiss)
[J].Chromosomal ploidy manipulation is one of the means to create excellent germplasm. Triploid fish could provide an ideal sterile model for searching of a underlying mechanism of abnormality in meiosis. The complete understanding of the coding and noncoding RNAs regulating sterility caused by meiosis abnormality is still not well understood. By high-throughput sequencing, we compared the expression profiles of gonadal mRNA, long non-coding RNA (lncRNA), and microRNA (miRNA) at three different developmental stages between the diploid (XX) and triploid (XXX) female rainbow trout. These stages were gonads before differentiation (65 days post fertilisation, dpf), at the beginning of morphological differences (180 dpf) and showing clear difference between diploids and triploids (600 dpf), respectively. A majority of differentially expressed (DE) RNAs were identified, and 22 DE mRNAs related to oocyte meiosis and homologous recombination were characterized. The predicted miRNA-mRNA/lncRNA networks of 3 developmental stages were constructed based on the target pairs of DE lncRNA-miRNA and DE mRNA-miRNA. According to the networks, meiosis-related gene of ccne1 was targeted by dre-miR-15a-5p_R + 1, and 6 targeted DE lncRNAs were identified. Also, qRT-PCR was performed to validate the credibility of the network. Overall, this study explored the potential interplay between coding and noncoding RNAs during the gonadal development of polyploid fish. The mRNA, lncRNA and miRNA screened in this study may be helpful to identify the functional elements regulating fertility of rainbow trout, which may provide reference for character improvement in aquaculture.Copyright © 2018. Published by Elsevier Inc.
Effects of letrozole and 17α-methyltestosterone on gonadal development in all-female triploid rainbow trout (Oncorhynchus mykiss)
[J].
Oxidative stress-related gene expression in diploid and triploid rainbow trout (Oncorhynchus mykiss) fed diets with organic and inorganic zinc
[J].
Effects of feeding level and sexual maturation on expression of genes regulating growth mechanisms in rainbow trout (Oncorhynchus mykiss)
[J].
DNA barcoding identification of commercialized seafood in South Brazil: a governmental regulatory forensic program
[J].
Development of real-time PCR assay for genetic identification of the mottled skate, Beringraja pulchra
[J].The mottled skate, Beringraja pulchra is one of the commercially important fishes in the market today. However, B. pulchra identification methods have not been well developed. The current study reports a novel real-time PCR method based on TaqMan technology developed for the genetic identification of B. pulchra. The mitochondrial cytochrome oxidase subunit 1 (COI) nucleotide sequences of 29 B. pulchra, 157 skates and rays reported in GenBank DNA database were comparatively analyzed and the COI sequences specific to B. pulchra was identified. Based on this information, a system of specific primers and Minor Groove Binding (MGB) TaqMan probe were designed. The assay successfully discriminated in 29 specimens of B. pulchra and 27 commercial samples with unknown species identity. For B. pulchra DNA, an average Threshold Cycle (Ct) value of 19.1±0.1 was obtained. Among 27 commercial samples, two samples showed average Ct values 19.1±0.0 and 26.7±0.1, respectively and were confirmed to be B. pulchra based on sequencing. The other samples tested showed undetectable or extremely weak signals for the target fragment, which was also consistent with the sequencing results. These results reveal that the method developed is a rapid and efficient tool to identify B. pulchra and might prevent fraud or mislabeling during the distribution of B. pulchra products. Copyright © 2015 Elsevier Ireland Ltd. All rights reserved.
A single-laboratory validated method for the generation of DNA barcodes for the identification of fish for regulatory compliance
[J].The U.S. Food and Drug Administration is responsible for ensuring that the nation's food supply is safe and accurately labeled. This task is particularly challenging in the case of seafood where a large variety of species are marketed, most of this commodity is imported, and processed product is difficult to identify using traditional morphological methods. Reliable species identification is critical for both foodborne illness investigations and for prevention of deceptive practices, such as those where species are intentionally mislabeled to circumvent import restrictions or for resale as species of higher value. New methods that allow accurate and rapid species identifications are needed, but any new methods to be used for regulatory compliance must be both standardized and adequately validated. "DNA barcoding" is a process by which species discriminations are achieved through the use of short, standardized gene fragments. For animals, a fragment (655 base pairs starting near the 5' end) of the cytochrome c oxidase subunit 1 mitochondrial gene has been shown to provide reliable species level discrimination in most cases. We provide here a protocol with single-laboratory validation for the generation of DNA barcodes suitable for the identification of seafood products, specifically fish, in a manner that is suitable for FDA regulatory use.
Gonadotropin-releasing hormone receptor (GnRHR) gene expression is differently modulated in gender types of the hermaphroditic fish Kryptolebias marmoratus by endocrine disrupting chemicals
[J].
Genomic diversity of type B3 bacteriophages of Caulobacter crescentus
[J].
MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets
[J].We present the latest version of the Molecular Evolutionary Genetics Analysis (Mega) software, which contains many sophisticated methods and tools for phylogenomics and phylomedicine. In this major upgrade, Mega has been optimized for use on 64-bit computing systems for analyzing larger datasets. Researchers can now explore and analyze tens of thousands of sequences in Mega The new version also provides an advanced wizard for building timetrees and includes a new functionality to automatically predict gene duplication events in gene family trees. The 64-bit Mega is made available in two interfaces: graphical and command line. The graphical user interface (GUI) is a native Microsoft Windows application that can also be used on Mac OS X. The command line Mega is available as native applications for Windows, Linux, and Mac OS X. They are intended for use in high-throughput and scripted analysis. Both versions are available from www.megasoftware.net free of charge.© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Spectral neighbor joining for reconstruction of latent tree models
[J].A common assumption in multiple scientific applications is that the distribution of observed data can be modeled by a latent tree graphical model. An important example is phylogenetics, where the tree models the evolutionary lineages of a set of observed organisms. Given a set of independent realizations of the random variables at the leaves of the tree, a key challenge is to infer the underlying tree topology. In this work we develop Spectral Neighbor Joining (SNJ), a novel method to recover the structure of latent tree graphical models. Given a matrix that contains a measure of similarity between all pairs of observed variables, SNJ computes a spectral measure of cohesion between groups of observed variables. We prove that SNJ is consistent, and derive a sufficient condition for correct tree recovery from an estimated similarity matrix. Combining this condition with a concentration of measure result on the similarity matrix, we bound the number of samples required to recover the tree with high probability. We illustrate via extensive simulations that in comparison to several other reconstruction methods, SNJ requires fewer samples to accurately recover trees with a large number of leaves or long edges.
A review of the application of molecular genetics for fisheries management and conservation of sharks and rays
[J].Since the first investigation 25 years ago, the application of genetic tools to address ecological and evolutionary questions in elasmobranch studies has greatly expanded. Major developments in genetic theory as well as in the availability, cost effectiveness and resolution of genetic markers were instrumental for particularly rapid progress over the last 10 years. Genetic studies of elasmobranchs are of direct importance and have application to fisheries management and conservation issues such as the definition of management units and identification of species from fins. In the future, increased application of the most recent and emerging technologies will enable accelerated genetic data production and the development of new markers at reduced costs, paving the way for a paradigm shift from gene to genome-scale research, and more focus on adaptive rather than just neutral variation. Current literature is reviewed in six fields of elasmobranch molecular genetics relevant to fisheries and conservation management (species identification, phylogeography, philopatry, genetic effective population size, molecular evolutionary rate and emerging methods). Where possible, examples from the Indo-Pacific region, which has been underrepresented in previous reviews, are emphasized within a global perspective.© 2012 The Authors. Journal of Fish Biology © 2012 The Fisheries Society of the British Isles.
Molecular identification methods of fish species: reassessment and possible applications
[J].
DNA barcodes for ecology,evolution,and conservation
[J].
DNA barcoding vs. morphological identification of larval fish and embryos in Lake Huron: advantages to a molecular approach
[J].
Fish product mislabeling identified in the Russian far east using DNA barcoding
[J].
Marketplace substitution of Atlantic salmon for Pacific salmon in Washington State detected by DNA barcoding
[J].
The current status of DNA barcoding technology for species identification in fish value chains
[J].
DNA barcode identification of fish products in Taiwan: government-commissioned authentication cases
[J].
Development of a DNA mini-barcoding protocol targeting COⅠfor the identification of elasmobranch species in shark cartilage pills
[J].
Universal mini COⅠbarcode for the identification of fish species in processed products
[J].
DNA barcoding revealed mislabeling and potential health concerns with roasted fish products sold across China
[J].75.5% of products were identified as species outside the expected family. Six products were identified as containing multiple species from distinct families. Species from distinct families were verified in products of same brand for six groups. Identification of potentially toxic pufferfish species highlighted health concerns.
Phylogeny of salmonids (Salmoniformes: Salmonidae) and its molecular dating: analysis of nuclear RAG1 gene
[J].
Evolution of the charrs, genus Salvelinus (Salmonidae). 1. origins and expansion of the species
[J].
Low mislabeling rates indicate marked improvements in Eusropean seafood market operations
[J].Over the span of a decade, genetic identification methods have progressively exposed the inadequacies of the seafood supply chain, revealing previously unrecognized levels of seafood fraud, raising awareness among the public, and serving as a warning to industry that malpractice will be detected. Here we present the outcome of the latest and largest multi‐species, transnational survey of fish labeling accuracy to date, which demonstrates an apparent sudden reduction of seafood mislabeling in Europe. We argue that recent efforts in legislation, governance, and outreach have had a positive impact on industry regulation. Coordinated, technology‐based, policy‐oriented actions can play a pivotal role in shaping a transparent, sustainable global seafood market and in bolstering healthier oceans.
DNA barcode identification of fish products from Guiyang markets in southwestern People’s Republic of China
[J].
Identifying introgressive hybridization in native populations of California golden trout based on molecular markers
[J].The California golden trout Oncorhynchus mykiss aguabonita is one of three subspecies within the rainbow trout–redband trout complex endemic to the Kern River basin and historically restricted to Golden Trout Creek and the South Fork Kern River. Past allozyme studies have indicated that native populations of California golden trout in the Golden Trout Creek drainage may have become introgressed with rainbow trout alleles through interaction with hybrids of rainbow trout and California golden trout stocked into nearby headwater lakes that are connected to tributaries in the drainage. We used six microsatellites and a minisatellite marker to estimate the genetic diversity and levels of introgression in approximately 700 California golden trout taken from 23 locations in Golden Trout Creek, its tributaries, and surrounding lakes. Indications of introgression were found in all but one of the sampled Golden Trout Creek drainage locations, the lowest average levels (0–8%) occurring in the lower reaches of Golden Trout Creek. The highest levels (12–17%) were in the Cottonwood Lakes populations that have been used as California golden trout broodstock by the California Department of Fish and Game. Evidence of introgression was also found in fish sampled from the upper reaches of the South Fork Kern River. This suggests that past and present stocking policies and hybridization with introduced rainbow trout currently threaten the genetic integrity of California golden trout populations across all of their native range.
Subspecies-informative SNP assays for evaluating introgression between native golden trout and introduced rainbow trout
[J].We characterize 20 single nucleotide polymorphism assays for evaluating hybridization between native golden trout subspecies (Oncorhynchus mykiss aguabonita and O. m. whitei) and introduced rainbow trout strains. These assays utilize the 5'-nuclease reaction, facilitating high-throughput genotyping of many individuals and making them useful in quantifying and monitoring introgression and potentially applicable to studies of other O. mykiss groups. Minor allele frequency differentials (δq) among native and introduced rainbow groups ranged from 0 to 1, with an average differential of 0.75 for both subspecies aguabonita and whitei relative to the hatchery rainbow trout strain.© 2009 The Authors. Journal compilation © 2009 Blackwell Publishing Ltd.
The physiology of triploid fish: current knowledge and comparisons with diploid fish
[J].
Expression of genes associated with fatty acid metabolism during maturation in diploid and triploid female rainbow trout
[J].