肠道微生物是指动物肠道中存在的数量繁多、组成结构复杂多样的微生物菌群。这些微生物菌群依靠宿主动物的肠道生活,同时帮助寄生宿主完成多项正常的生理生化功能。目前,有研究表明鱼类肠道中存在丰富的微生物菌群[1],也发现大多数鱼类的疾病发生和免疫功能与其肠道微生物菌群有着一定的相关性[2],且机体的肠道动态平衡状态是动物生长发育过程中不可或缺的一个部分。在鱼体中,保持鱼体肠道中微生物菌群的稳态可有效地帮助鱼类在消化吸收、免疫系统、机体健康等方面发挥作用[3]。当鱼体遭受致病性细菌侵袭之后,其肠道菌群的平衡便会产生变化,进而导致整个鱼体免疫功能异常甚至失调[4],如鱼体肠道内病原菌或致病菌的异常增多,进而可能导致鱼体生病、损伤甚至死亡[5]。此外,鱼类肠道微生物还肩负着许多鱼体的功能性重任,如鱼类肠道微生物菌群可以合成多种维生素,作为辅酶参与和保护细胞、组织及器官的正常生理生化过程[6]。
青藏高原地区包括青海和西藏,作为一个特殊的环境地带,该地区具有高海拔、低氧、强紫外线辐射和冬季低温等气候特点[7]。同时,复杂多样的特殊地域条件和高寒气候造就了其所生存鱼类的独特性,形成了该区域鱼类个体生长繁殖速度迟缓、性成熟发育时间长等生物学特点[8],鱼类种群一旦遭到破坏,将极难恢复[9]。此外,青藏高原地区在历史上的整体水生态系统和演化时间都比较短,导致其水生态系统中的物种种间竞争相对较为宽松,同时还存在着许多水生态系统空余生态位[10],当外来鱼入侵时,高原土著鱼类的生态位空间极易被占领。如能通过调节肠道微生物这一手段对高原鱼类的养护进行探索,了解鱼类肠道微生态结构,对高原鱼类养护工作及西藏鱼类生态系统保护意义深远。因此,本文对维持鱼体肠道微生态平衡稳定的重要因素及常见鱼类肠道微生物菌群的主要研究技术手段进行介绍,为未来西藏鱼类养护提供基础资料。
1 西藏土著鱼类资源现状
1.1 西藏土著鱼类资源概况
近20年来,各种因素汇集使西藏的渔业生态变得异常脆弱,据研究人员统计,在当前西藏旅游经济和社会快速发展的新时期,青藏高原所有的 162 种鱼类[11-12]中,处于极危(CR)、濒危(EN)、易危(VU)和野外绝灭(EW)的鱼类[13]有35 种,占文献已记载的超过20%[14]。自2014年以来,刘海平等[15]对雅鲁藏布江各江段鱼类分布情况进行了调查分析,雅鲁藏布江流域共有28种鱼类,隶属2目4科(亚科),主要裂腹鱼类有6种,即拉萨裂腹鱼(Racoma waltoni)、巨须裂腹鱼(Schizothorax macropogon)、异齿裂腹鱼(Schizothorax oconnori)、尖裸鲤(Oxygymnocypris stewarti)、双须叶须鱼(Ptychobarbus dipogon)以及拉萨裸裂尻鱼(Schizopygopsis younghusbandi)。西藏地区鱼类60种、13亚种,分属于3目、5科、4亚科、22属,其中鲑科1种,占1.4%;鲤科42种,占58.0%;裸吻鱼科1种,占1.4%;鳅科16种,占22.0%; 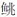 科13种,占18.0%。以上裂腹鱼类和
科13种,占18.0%。以上裂腹鱼类和 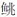 科鱼类居多,极危、濒危、易危、近危的鱼类有23种,占近36.0%[15]。从以上的数据均可看出,目前西藏鱼类资源和种类都处于一个相对危险的状态,因此进行西藏土著鱼类养护、资源开发和利用尤为紧迫与重要。
科鱼类居多,极危、濒危、易危、近危的鱼类有23种,占近36.0%[15]。从以上的数据均可看出,目前西藏鱼类资源和种类都处于一个相对危险的状态,因此进行西藏土著鱼类养护、资源开发和利用尤为紧迫与重要。
此外,随着西藏地区外来人口增多和旅游业的大力发展,西藏各地区对于水产品的需求日益增加,进而导致过度捕捞和外来鱼类的入侵等,雅鲁藏布江主要裂腹鱼类的种群数量开始大幅度下降,特别是大型鱼类个体正在逐渐减少。雅鲁藏布江主要裂腹鱼类资源现状也已敲响了警钟,比如分布于雅鲁藏布江中游主要的6种裂腹鱼类,仅有拉萨裂腹鱼[16]和异齿裂腹鱼[17]目前的利用程度不高,自然资源还可持续发展;其他4种裂腹鱼的种群现状已然达不到原始的自然生长状态,特别是尖裸鲤的种群资源已被人类过度利用[18],自然种群结构已被破坏;双须叶须鱼[19]、巨须裂腹鱼[20]种群中的雌性群体大多为过度捕捞状态,雄性群体处于自然死亡率较高状态;拉萨裸裂尻鱼中的雌性个体也已经处于过度捕捞状态,繁殖潜力比大大降低,据数据统计均低于下限参考点F25%,而雄性个体已经处于完全开发状态,繁殖潜力比相对雌性群体高,接近目标参考点F40%[21-22]。随着国家大力发展西部,西部大开发战略在西藏地区的实施卓有成效,西藏地区经济发展持续稳步上升,但该地区的生态资源保护及可持续利用发展也不容忽视[23],就此现状来看,西藏地区鱼类及雅鲁藏布江主要鱼类的科学养护研究迫在眉睫。因此,必须加快推动雅鲁藏布江主要鱼类的养护工作[8]。
1.2 西藏土著鱼类的特点
西藏土著鱼类在高原上长期生存,其生长繁殖已逐步适应高原环境,因而表现出与平原鱼类有所不同的生物学特征。如长期受高原高寒、低温、少氧的水体环境条件影响,食物相对较匮乏,因此西藏土著鱼类的生物学特性具有生长时间长、性成熟时间晚、繁殖能力低等特点[24]。同时,因为青藏高原地区水温一般较低,所以在这样的水体温度中生活的鱼类大多为西藏当地的特有品种。其种群和群落结构一旦遭到破坏,整个种群和群落要重新恢复,所需时间极长、难度也极大。例如,色林错裸鲤(Gymnocypris selincuoensis)要达到个体性发育成熟、具有繁殖能力且体质量在500 g左右的水平,整个生长时间就至少需要10年。
相关研究表明,不同食性鱼类肠道微生物菌群的组成各有不同;相同食性、不同种类的鱼类肠道微生物菌群也有所不同,这为食性相异性的雅鲁藏布江特有鱼类的研究工作开拓了新的思路,雅鲁藏布江特有鱼类分布区域及食性详见表1。
表1 雅鲁藏布江特有鱼类分布区域及食性
Tab.1
| 雅鲁藏布江鱼类 Fish of Brahmaputra River | 雅鲁藏布江分布区域 Distribution area in the Brahmaputra River | 食性 Feeding habits | |||
|---|---|---|---|---|---|
| 上游 Upstream | 中上游 Mid-to-up | 中下游 Lower reaches | 下游 Downstream | ||
| ●鲤形目 Cypriniformes | |||||
| ●●条鳅亚科 Noemacheilinae | |||||
| ●●●条鳅属 Nemacheilus | |||||
| ●●●●浅棕条鳅 Nemacheilus subfuscus McClelland | + | 待研究 | |||
| ●●●高原鳅属 Triplophysa | |||||
| ●●●●东方高原鳅 Triplophysa orientalis Herzenstein | + | + | + | 待研究 | |
| ●●●●西藏高原鳅 Triplophysa tibetana Regan | + | + | + | 待研究 | |
| ●●●●斯氏高原鳅 Triplophysa stoliczkai Steindachner | + | + | + | 待研究 | |
| ●●●●细尾高原鳅 Triplophysa stenura Herzenstein | + | + | + | 待研究 | |
| ●●鲃亚科 Barbinae | |||||
| ●●●四须鲃属 Barbodes | |||||
| ●●●●墨脱四须鲃 Barbodes hexagonolepis McClelland | + | 杂食性 | |||
| ●●●墨头鱼属 Garra | |||||
| ●●●●西藏墨头鱼 Garra kempi Hora | + | 待研究 | |||
| ●●野鲮亚科 Labeoninae | |||||
| ●●●华鲮属 Bangana | |||||
| ●●●●墨脱华鲮 Sinilabeo dero Hamilton | + | 杂食性 | |||
| ●●裂腹鱼亚科 Schizothoracinae | |||||
| ●●●裂腹鱼属Schizothorax | |||||
| ●●●●墨脱裂腹鱼 Schizothorax molesworthi Chaudhuri | + | 杂食性 | |||
| ●●●●异齿裂腹鱼 Schizothorax oconnori Lloyd | + | + | + | 植食性 | |
| ●●●●弧唇裂腹鱼 Schizothorax curvilabiatus Wu et Tsao | + | 杂食性 | |||
| ●●●●拉萨裂腹鱼 Schizothorax waltoni Regan | + | + | + | 杂食性 | |
| ●●●●巨须裂腹鱼 Schizothorax macropogon Regan | + | + | + | 杂食性 | |
| ●●●叶须鱼属 Ptychobarbus | |||||
| ●●●●双须叶须鱼 Ptychobarbus dipogon Regan | + | + | + | 杂食性 | |
| ●●●尖裸鲤属 Oxygymnocypris | |||||
| ●●●●尖裸鲤 Oxygymnocypris stewartii Lloyd | + | + | 肉食性 | ||
| ●●●裸裂尻鱼属 Schizopygopsis | |||||
| ●●●●拉萨裸裂尻鱼 Schizopygopsis younghusbandi Regan | + | + | + | 植食性 | |
| ●●●裸鲤属 Gymnocypris | |||||
| ●●●●软刺裸鲤 Gymnocypris dobula Günther | + | 待研究 | |||
| ●●●●兰格湖裸鲤 Gymnocypris chui Tchang,et al | + | 待研究 | |||
| ●●●●硬刺裸鲤 Gymnocypris firmispinatus Wu et Wu | + | 待研究 | |||
| ●●裸吻鱼科 Psilorhynchidae | |||||
| ●●●裸吻鱼属 Psilorhynchu | |||||
| ●●●●平鳍裸吻鱼 Psilorhynchus homaloptera Hora et Mukerji | + | 待研究 | |||
| ●鲇形目 Siluriformes | |||||
●● 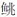 科 Sisoridae 科 Sisoridae | |||||
●●●纵纹  属 Glyptothorax 属 Glyptothorax | |||||
●●●●墨脱纹胸  Glyptothorax annandalei Hora Glyptothorax annandalei Hora | + | 肉食性 | |||
●●●●细体纹胸  Glyptothorax gracilis Günther Glyptothorax gracilis Günther | + | 待研究 | |||
●●●褶  属 Pseudecheneis 属 Pseudecheneis | |||||
●●●●黄斑褶  Pseudecheneis sulcatus McClelland Pseudecheneis sulcatus McClelland | + | + | 待研究 | ||
●●●平唇  属 Parachiloglanis 属 Parachiloglanis | |||||
●●●●平唇  Parachiloglanis hodgarti Hora Parachiloglanis hodgarti Hora | + | 待研究 | |||
●●●原  属 Glyptosternon 属 Glyptosternon | |||||
●●●●黑斑原  Glyptosternon maculatum Regan Glyptosternon maculatum Regan | + | + | 肉食性 | ||
●●●  属 Pareuchiloglanis 属 Pareuchiloglanis | |||||
●●●●扁头  Pareeuchiloglanis kamengensis Jayaram Pareeuchiloglanis kamengensis Jayaram | + | 待研究 | |||
●●●凿齿  属 Glaridoglanis 属 Glaridoglanis | |||||
●●●●凿齿  Glaridoglanis andersoni Day Glaridoglanis andersoni Day | + | ||||
●●●  属 Exostoma 属 Exostoma | |||||
●●●●藏  Exostoma labiatum McClelland Exostoma labiatum McClelland | + | 待研究 | |||
注:●为目,●●为科,●●●为属,●●●●为种;分布区域中“+”表示该鱼类在此区域中分布。
Notes: ● meant the order, ●● meant the family, ●●● meant the genus, and ●●●● meant species; “+”in the distribution area indicated that the fish was distributed in this area.
1.3 西藏鱼类资源养护存在的问题
近年来,由于国家政策和西藏自治区的发展规划,西藏的经济和旅游业正在快速发展,西藏地区外来人口日渐增多,从而对西藏野生鱼类和养殖鱼类的广泛需求日益增长,导致现在西藏本土地区鱼类的捕捞越来越严重。由于酷渔滥捕、人工移植,西藏土著鱼类的栖息地正在不断减少,造成其种群数量不断下降,自然条件下的鱼类资源被严重破坏,自然资源日益短缺。此外,受城市水电工程建设和居民生活排放污水等影响,西藏土著鱼类的栖息环境正在不断改变[9]。水环境的改变导致鱼类患病的现象越来越多,造成西藏土著鱼类种群数量的急速锐减,从而使西藏鱼类资源受损。例如在 2009年,雅鲁藏布江中下游流域段曾经出现了大量的死鱼,损失了大量的自然渔业资源,对西藏渔业生态系统造成了巨大的冲击[27]。
2 鱼类肠道微生物的种类与研究方法
2.1 鱼类肠道微生物的种类
在鱼类的肠道微生物中,其组成菌类大多为细菌和真菌等[27]。在这些组成成分中,细菌为主要类群,包括变形菌门(Proteobacteria)、梭杆菌门(Fusobacteria)、厚壁菌门(Firmicutes)、拟杆菌门(Bacteroidetes)、放线菌门(Actinobacteria)和疣微菌门(Verrucomicrobia)等细菌[28],其中鱼类肠道微生物有3种相同的菌门,分别为变形菌门、厚壁菌门和拟杆菌门,且占到很高的比例,表明它们可能对鱼类肠道有着重要的作用和影响[29]。针对不同食性淡水鱼类的肠道微生物群落,研究结果(表2)发现其肠道微生物菌群在组成和种类上均存在着差异[30]。
表2 常见淡水鱼类肠道微生物菌群组成
Tab.2
| 食性 Feeding habits | 食物 Food | 常见淡水鱼 Common freshwater fish | 肠道微生物菌群 Gut microbiota |
|---|---|---|---|
| 植食性 Phytovorous | 水草 丝状藻类 水中的其 他植物 | 鳊(Parabramis pekinensis) 草鱼(Ctenopharyngodon idellus) | 气单孢菌、假单胞菌、拟杆菌门[31]、变形菌门、厚壁菌门、放线菌门[32]、梭杆菌属[33]、链球菌属、哈尼亚菌属、致病杆菌属、柠檬酸菌属和葡萄球菌属[34]、嗜酸乳杆菌、双歧杆菌[35] |
| 浮游生物 | 鲢(Hypophthalmichthys molitrix) | 哈尼亚菌属、致病杆菌属、气单孢菌属、柠檬酸杆菌属、假单孢杆菌属、拉恩氏菌属、链球菌属[34]、嗜酸乳杆菌、双歧杆菌、变形菌门、拟杆菌门、厚壁菌门、疣微菌门、蓝细菌门[36] | |
| 鳙(Aristichthys nobilis) | 拟杆菌门、哈尼亚菌属、致病杆菌属、气单孢菌属、柠檬酸杆菌属、假单孢杆菌属、拉恩氏菌属、链球菌属[34]、嗜酸乳杆菌、双歧杆菌[35]、变形菌门、梭杆菌门、厚壁菌门、疣微菌门[37]、蓝细菌门[36]、放线菌门、浮霉菌门 | ||
| 鲤(Cyprinus carpio) | 弧菌、气单胞菌[35]、变形菌门、拟杆菌门、梭杆菌门、浮霉菌门、链球菌、哈尼亚菌属、致病杆菌属、柠檬酸杆菌属[38] | ||
| 杂食性 Omnivorous | 动物性食物 植物性食物 | 鲫(Carassius auratus) | 变形菌门、厚壁菌门、梭杆菌门、拟杆菌门[33]、弧菌、气单孢菌[35]、蓝藻细菌、放线菌门[39] |
| 斑马鱼(Danio rerio) | 梭菌属、蓝细菌、变形菌门、放线菌门[40] | ||
| 肉食性 Carnivorous | 捕捉其他 鱼类为食 | 青鱼 (Mylopharyngodon piceus) 翘嘴红鲌 (Erythroculter ilishaeformis) 乌鳢(Channa argus) | 链球菌属、哈夫尼亚菌属、致病杆菌属、气单孢菌属柠檬酸菌属、葡萄球菌属[34]、厌氧芽孢杆菌[35] |
2.2 鱼类肠道微生物的研究方法
表3 鱼类肠道微生物研究相关手段方法
Tab.3
| 手段和方法 Means and methods | 内容 Content | 优点 Merit | 缺点 Shortcoming | 相关文献 Related literature | |||||
|---|---|---|---|---|---|---|---|---|---|
| Illumina Miseq 平台、16S rRNA的V3~V4区测序、OTUs | 运用Illumina Miseq平台对肠道微生物16S rRNA的V3~V4区进行了测序,统计样品肠道微生物的操作分类单元(OTUs)数量,分析物种组成、丰度及Alpha多样性,并预测肠道微生物的功能 | 成本低 | 无法获得功能基因,群落信息有限 | 刘妮等[43] | |||||
| DNA试剂盒 | 使用试剂盒法提取肠道菌群总 DNA,电泳结果显示,样品DNA 条带明亮,无降解现象;通过设计的细菌通用引物,对其16S rDNA 基因进行 PCR扩增,得到较清晰的图谱,条带整齐 | 试剂盒应用方便、快捷 | 价格昂贵,在处理大量样品时成本较高 | 汪明等[44] | |||||
| PCR-DGGE 指纹分析技术 | 使用PCR-DGGE指纹分析技术对室内饲养的斑点叉尾  (Ictalurus punctatus)、银鲫(Carassius auratus gibelio)和异育银鲫(Carassiusauratus gibelio)3种鱼类的肠道微生物群落组成进行研究 (Ictalurus punctatus)、银鲫(Carassius auratus gibelio)和异育银鲫(Carassiusauratus gibelio)3种鱼类的肠道微生物群落组成进行研究 | 肠道菌群的信息涵盖率高 | 易受DNA提取、PCR 扩增和电泳条件等多种因素的影响,无法全面准确展示肠道菌群结构 | 李学梅等[45] | |||||
| 变性梯度凝胶电泳(DGGE) | 利用PCR-DGGE技术对鳜受精卵、出膜仔鱼、仔稚鱼和其开口饵料鲢仔稚鱼的肠道微生物群落结构组成和多样性进行分析 | 肠道菌群的信息涵盖率高 | 易受DNA提取、PCR 扩增和电泳条件等多种因素的影响,无法全面准确展示肠道菌群结构 | 夏耘等[46] | |||||
| 末端限制性片段长度多态性分析(T-RFLP) | 分析对虾肠道正常菌群的营养、消化、免疫和屏障等生理功能,及对虾肠道菌群结构与组成的影响因素 | 肠道菌群的信息涵盖率高 | 无法全面准确展示肠道菌群结构,难以精确地归类到种属 | 张家松等[47] | |||||
| 高通量测序技术、宏基因组学测序技术 | 采用宏基因组学测序技术和生物信息学分析手段,构建了草鱼、鲫、鲢和鳙4种鲤科鱼类的肠道内含物等12个样品的16S rDNA测序克隆文库,分析其肠道微生物的菌落组成和多样性 | 鉴定深度在种水平,检测全基因组DNA序列,可分析功能基因组和样品间基因差异、研究物种间代谢网络等 | 研究成本高 | 李建柱等[48] | |||||
| 16S rRNA基因V3~V4可变区、Illumina NovaSeq平台高通量测序、定量试剂盒 | 采用高通量测序技术分析丽水市千峡湖翘嘴鲌(Culter alburnus Basilewsky)、达氏鲌(Huso dauricus)和红鳍原鲌(Chanodichthys erythropterus)3种肉食性鱼类的肠道微生物群落结构 | 成本低,可分析微生物群结构,反映各种微生物菌群相对比例和数量,具有传统培养法难以比拟的优势 | 无法获得功能基因,群落信息有限 | 肖善势等[49] | |||||
| 生物纯培养 技术 | 分别从虾和鱼的肠道中分离出53个乳酸菌(Lactic acid bacteria,LAB)菌株进行考察这些菌株的抗菌和黏附活性 | 可以具体检测到种属,容易分类 | 培养要求高;人为培养无法满足需求,也无法准确得到需要的产物。培养条件较局限,检测出菌群种类少且存在较大偏差 | Sha Y J 等[50-51] | |||||
| 无菌斑马鱼 模型 | 开发了在幼年后期生产和养殖无菌斑马鱼的方法。对受精后6 d,无菌和常规饲养的斑马鱼消化道中基因表达的DNA微阵列比较揭示了212个由微生物群调节的基因 | 无菌斑马鱼繁殖能力强、饲养成本低、无菌操作便捷等,有助于研究者探讨微生物对宿主的营养、免疫、运动、神经发育等方面的调节作用 | 接种条件多种多样,对最佳接种条件仍不清楚,极大地限制了无菌动物模型的使用,降低了不同研究结果之间的可比性 | Rawls J F 等[52] | |||||
| 无菌斑马鱼 模型 | 使用2种斑马鱼模型(无菌和抗生素处理的斑马鱼)来确定肠道微生物菌群在脂质代谢中的作用,常规和无菌的斑马鱼幼虫用蛋黄喂养,检测肠上皮中脂质液滴的存在 | 无菌斑马鱼繁殖能力强、饲养成本低、无菌操作便捷等,有助于研究者探讨微生物对宿主的营养、免疫、运动、神经发育等方面的调节作用 | 接种条件多种多样,对最佳接种条件仍不清楚,极大地限制了无菌动物模型的使用,降低了不同研究结果之间的可比性 | Sheng Y 等[53] | |||||
| 粪菌移植技术 | 在有氧条件下从3只成年CONV-R Swiss-Webster雌性小鼠中收集盲肠内容物,在PBS中稀释1∶1 200,并直接(1∶100稀释)加入含有3 dpf GF斑马鱼的GZM[最终密度:102 CFU/mL(有氧培养);103 CFU/mL(厌氧培养)],在BHI血琼脂上28 ℃下孵育2 d | 方便可行,经济费用比较低 | 很难保证粪菌的有效性及稳定性 | Rawls J F 等[54] | |||||
2.3 鱼类肠道微生物的作用
正常鱼类肠道中的微生物菌群能够合成产生各种各样的消化酶,如纤维素酶、脂肪酶等,这些酶都是鱼类营养吸收的关键因素,因此,肠道微生物菌群对鱼类的生长发育具有重要影响[55]。此外,研究还发现正常的鱼类肠道微生物菌群可以合成维生素,如维生素B1、维生素B12和泛酸等,且在不同食性的鱼类肠道中合成的也不同,如植食性鱼类肠道中合成木聚糖分解菌、杂食性鱼类消化道内生长着甲壳质分解菌[30]。国外早期研究表明,鱼类肠道内的微生物在合成多种维生素时,被报道的合成最多、最常见的是维生素B12,且其被合成主要依靠厌氧型微生物[56];同时在其他的研究中也有发现,在多种鱼类肠道中通过分离而获得的专性厌氧型细菌能够分泌出鱼体消化吸收所需要的多种消化酶[57]。这说明厌氧型细菌可作为鱼体分泌多种消化酶的主要微生物菌种,也表明了鱼类肠道微生物群落的组成结构在鱼体的营养补给、食物消化和吸收方面有着重要作用。
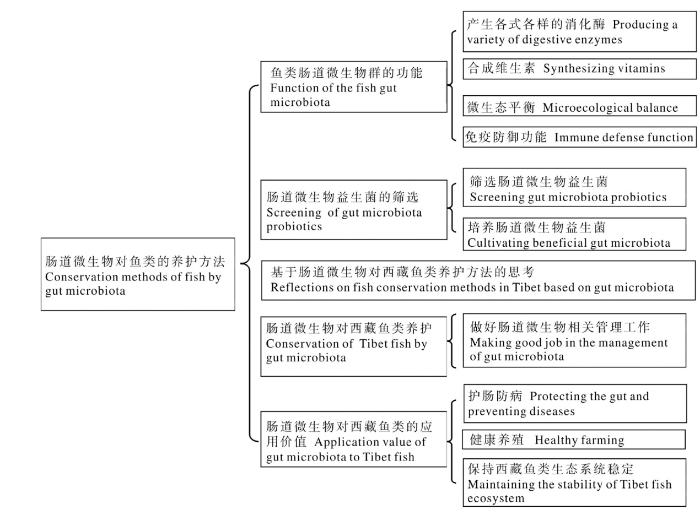
3 利用肠道微生物探索西藏土著鱼类资源养护方法
图1
3.1 筛选西藏土著鱼类肠道益生菌
水产动物肠道中的微生物菌群主要包括好氧细菌、兼性厌氧菌和专性厌氧菌[67]。不同的养殖环境和饲料会改变肠道微生物群落的结构,而其与水产动物疾病发生有着十分重要的关联,同时肠道微生物群落的组成受养殖环境微生物群落影响,进而影响水产动物健康[68]。因此,肠道微生物菌群的组成不仅可以反映宿主健康状态,还反映了水体环境情况。在水产养殖方面,乳酸菌被广泛认为是一种对所有动物肠道均有益的菌株,可在投喂生物或环境中产生抗菌肽、蛋白质等物质。在大环境中投放乳酸菌具有降低环境pH值的功能,这是因为环境中不耐酸的腐败菌和致病菌因环境条件改变而被抑制繁殖[69]。因此,乳酸菌可以调控生物体肠道功能,如通过合成机体所需氨基酸、维生素等生长发育营养物质,促进机体生长和营养代谢;通过刺激机体免疫系统的调节来提高免疫力[69]。在对西藏土著鱼类的研究中,可以尝试以本土健康鱼体肠道中的细菌为候选菌,筛选出具有益生效果的菌株,再将这些菌株制备成复合益生菌剂,应用于土著鱼类饲喂试验,研究其对西藏土著鱼类体生长、免疫功能和胃肠道菌群组成的影响。
3.2 培养有益西藏土著鱼类肠道的微生物
鱼类肠道中存在着一个平稳的微生态环境,在正常鱼类肠道微生物群落中,整个稳态环境一般不会被破坏,还兼具辅助机体营养物质的消化吸收和控制机体寄生虫、有害菌等生长的作用。在鱼体的整个肠道微生态环境中,微生物与鱼体处于一个互利共生的平衡中,鱼体为寄生微生物菌群提供生长繁殖条件,微生物菌群为鱼体提供营养物质和发挥多种有益作用,从而形成一个统一的整体[70]。目前,对于西藏渔业资源来说,开展渔业养护的工作十分重要,而在这其中积累人工繁殖经验和推广繁育技术更为重要。但是目前西藏土著鱼类人工养殖的繁育技术还处于不稳定状态,其种质资源保存和开发也存在一些问题,包括市场推广风险和某些繁育技术瓶颈暂时无法突破,这成为了西藏土著鱼类人工繁育的卡脖子问题[8]。
在进行西藏土著鱼类人工繁育过程中,所养殖的每一种鱼不仅养育了鱼本身,也养育了包含其中的数万亿种微生物。因此,要想解决鱼病、饲料等问题,就得培养和维持健康的鱼类肠道菌群,从根本上了解西藏鱼类肠道微生物群落的多样性和组成特征及其对鱼类生长和健康的影响。例如,陈美群等[71]研究发现,病变死亡后尖裸鲤的肠道中 Acinetobacter sp.、Flavobacterium sp.、Vagococcus sp.、Carnobacterium sp.、Bacillus sp.和 Malassezia sp.等 6 种优势微生物菌群减少,而Pseudomonas sp.、Tremellales 和 Agaricomycetes 等3种微生物菌群增加,此外研究中还发现有 Pseudoperkinsidae 等病原动物存在于尖裸鲤皮肤和肠道中。由此可知,在尖裸鲤的人工养殖中适当提高Acinetobacter sp.等优势微生物菌的占比、控制Pseudoperkinsidae等病原动物在人工养殖环境中的存在,可为尖裸鲤人工养殖的疾病防控提供参考。Pan Y Z等[72]研究发现,噬铜菌是黑斑原肠道的特性核心微生物。因此,在人工饲养黑斑原时,可以增加其饲料中铜离子的含量,找到适宜浓度,从而达到既能满足黑斑原对饮食中铜离子的需求,又能抑制病原菌的生长,使黑斑原在人工养殖条件下健康生长。鱼类的先天免疫、适应性免疫等功能可以通过肠道中益生菌调节平衡,益生菌通过改变鱼类肠道中的微生物结构,建立肠道免疫和机体免疫,从而形成微生态平衡系统[73-74]。因此在养殖过程中,人工培养出西藏土著鱼类肠道中特有的益生菌,对维系西藏土著鱼类肠道中微生物菌群微生态平衡、抑制鱼体内有害菌的生长、改善鱼体健康和防控鱼病十分有效。但一切都要基于养殖中鱼群实际情况而定,对投喂量、投喂品种等都需要提前进行实验来确定最佳浓度配比和合适需求量,杜绝和减少鱼体肠道因饲料问题而造成的刺激和失衡,或因鱼体肠道负担重而产生炎症和其他疾病,从而达到有效提高鱼体肠道中有益微生物的比例和各种消化分解酶的合成等,改善鱼体肠道功能、提高鱼体健康水平。
3.3 做好西藏土著鱼类肠道微生物相关管理工作
鱼类在抵御外来病原菌时的首个屏障是肠道,因此要预防鱼病,首先要养护好鱼肠道。渔业人工养殖中大多使用密集型集约化养殖模式,这不仅会增加鱼群中各鱼体微生物生态失衡的可能性,还会加大鱼体感染病原和出现炎症的风险[75]。因此,利用肠道微生物调控的方法来提升鱼类繁育技术对西藏土著鱼类的可持续发展至关重要。目前,肠道微生物菌群调控机体健康的方法主要集中在人类疾病预防与治疗研究上,而在水生生物生产繁育等方面的研究少有报道[76]。在鱼类养护中,把肠道微生物结构、功能等与西藏土著鱼类人工繁育技术相联系,对于发展西藏土著鱼类生产实践工作十分有益。首先,确定引起疾病的单一致病微生物和鉴定其余的微生物群落,以确定它们对病原体产生了某种抗体,再开发出新的饲料,用于预防鱼类疾病,减少鱼类因疾病而造成的损失,促进西藏土著鱼类的健康养殖等。其次,现在对鱼类体内微生物菌群的产生时间、产生地点及产生方式均不确定,因此需要了解上述信息,以便最大限度地发挥这一新领域在西藏水产养殖方面的潜力。在对西藏土著鱼类的人工繁殖养护时,科学管理、养好水质护好肠,从而达到鱼体健康、繁殖量高、饵料系数低、死鱼量少的状态,才能真正实现西藏土著鱼类的健康养殖。
由于西藏土著鱼类栖息地不同,它们的生活习性及对周边环境的适应性也不同,因此每种鱼原生水域的生态环境、繁殖习性等对该种鱼苗的培育和成鱼的饲养管理都有其自身的特性。且在人工养殖条件下,环境的胁迫、饲料配方不佳、投喂方式不良等因素都容易引起养殖动物胃肠道的非健康状态。比如营底栖攀爬的科鱼类,需根据其生活和繁殖习性,采用鹅卵石铺垫、溶解氧充足、控制水流流速及温度的方法,模仿自然生活环境,提高其适应性,并采用活鱼和血虫作为混合鱼料的方法进行饲养。另外,还得多注意观察养殖池内鱼的活动情况,做到早发现早治疗。在实际养殖中,鱼体肠道发生疾病而用药的原因大多是由高密度养殖、水质恶化、饲料等所致。因此,考虑到环境条件和鱼体状况,治疗鱼病可选用受水体透明度、pH值等影响小的药剂,避免因使用刺激性药物而对鱼体造成影响。同时注意维护养殖水体环境的卫生,定期进行水环境改良,并不定期消毒,维护水体健康。因地制宜,因特制方,才能做好西藏土著鱼类的人工养护工作。
4 总结
随着鱼类肠道微生物菌群研究的不断深入,以及三代测序技术的快速发展应用,关于鱼类肠道微生物群落的研究也正在逐步深入。深入地了解西藏土著鱼类肠道微生物群落结构,对西藏地区鱼类健康发展起着重要的作用。目前西藏地区的水产业发展较为落后,与国内其他地区相比,其仍处于一个较低的发展水平。其中,关于西藏土著鱼类的研究在历史上很长一段时间大多处于空白,有的科学研究也是在近些年才逐步开始其发展进程。因此,以肠道微生物视角开展西藏土著鱼类的研究,对其资源养护具有重要意义。
参考文献
鱼类肠道微生物多样性及其与环境因子关系的研究进展
[J].早期认为鱼类肠道中存在寥寥可数、结构简单的微生物群落,随着鱼类肠道微生态学的发展,人们逐渐认识到鱼类肠道中存在着种类繁多、数量庞大的微生物群落。复杂的生活环境、不同的发育阶段、多变的食物来源等因素都会导致鱼类肠道微生物多样性及结构发生变化。鱼类肠道微生物与栖息环境所形成的微生态系统在保持动态平衡时,能够维持宿主的肠道健康。大量研究显示稳态中的微生态系统能够调节肠道微生物群落的平衡、改善肠道微生物群落的组成,从而对鱼类生长发育、营养代谢、免疫调控等过程具有促进作用。本文综述了鱼类肠道微生物的研究方法、多样性和功能,总结了温度、盐度及水体饵料等环境因子对鱼类肠道微生物的影响,以期为今后鱼类肠道微生物的研究发展提供启发和参考。
Gut microbiota in the pathogenesis of inflammatory bowel disease
[J].Inflammatory bowel disease (IBD), including ulcerative colitis and Crohn's disease, is a chronic and relapsing inflammatory disorder of the intestine. Although its incidence is increasing globally, the precise etiology remains unclear and a cure for IBD has yet to be discovered. The most accepted hypothesis of IBD pathogenesis is that complex interactions between genetics, environmental factors, and the host immune system lead to aberrant immune responses and chronic intestinal inflammation. The human gut harbors a complex and abundant aggregation of microbes, collectively referred to as the gut microbiota. The gut microbiota has physiological functions associated with nutrition, the immune system, and defense of the host. Recent advances in next-generation sequencing technology have identified alteration of the composition and function of the gut microbiota, which is referred to as dysbiosis, in IBD. Clinical and experimental data suggest dysbiosis may play a pivotal role in the pathogenesis of IBD. This review is focused on the physiological function of the gut microbiota and the association between the gut microbiota and pathogenesis in IBD. In addition, we review the therapeutic options for manipulating the altered gut microbiota, such as probiotics and fecal microbiota transplantation.
青藏高原的外来鱼类
[EB/OL].(2017-06-17)[2023-02-15]. http://www.ihb.cas.cn/kxcb_1/kxcb/202103/t20210311_5973375.html.
拉萨市拉鲁湿地鱼类现状与保护
[J].拉鲁湿地是西藏拉萨河流域重要的湿地之一。近年来,由于人类活动的影响,拉鲁湿地生态系统发生了较大的变化。本文于2010年4月、6月及2011年4月,通过刺网和地笼对拉鲁湿地的鱼类现状进行了调查。结果表明,拉鲁湿地现有7 种外来鱼类:鲫Carassius auratus (Linnaeus)、鲤Cyprinus caupio Linnaeus、草鱼Ctenopharyngodon idellus (Cuvier et Valenciennes)、麦穗鱼Pseudorasbora parva (Temminck et Schlegel)、泥鳅Misgurnus anguillicaudatus (Cantor)、鲇Silurus asotus Linnaeus、黄黝鱼Hypseleotris swinhonis (Gunther)和5种土著鱼类:尖裸鲤Oxygymnocypris stewartii (Lloyd)、拉萨裸裂尻鱼Schizopygopsis younghusbandi younghusbandi Regan、异尾高原鳅Triplophysa stewartii(Hora)、细尾高原鳅T. stenura (Herzenstein)、西藏高原鳅T. tibetana (Regan)。外来的麦穗鱼和鲫为绝对优势种,而5种土著鱼类的数量极少、几近灭绝。不均衡的群落结构导致拉鲁湿地的鱼类多样性极低。在分布上,麦穗鱼和鲫几乎遍布整个湿地,而土著鱼类仅分布于小部分水域中。当前,拉鲁湿地的鱼类群落已散失其独特性,而转变为以外来鱼类为主体、部分土著鱼类残存的格局。今后,改善拉鲁湿地的水质、加强群众对外来鱼类识别的能力和危害的认识、清除外来鱼类等措施对于保护拉鲁湿地的鱼类及其生态系统将有重要的意义。
Dietary Lactobacillus plantarum ST-Ⅲ alleviates the toxic effects of triclosan onzebrafish (Danio rerio) via gut microbiota modulation
[J].
Teleost intestinal immunology
[J].
Obligate anaerobic bacteria in the gastrointestinal microflora of the grass carp (Ctenopharyngodon idella), goldfish (Carassius auratus), and rainbow trout (Salmo gairdneri)
[J].
Composition, diversity, and origin of the bacterial community in grass carp intestine
[J].
Comparative analysis of the intestinal bacterial communities in different species of carp by pyrosequencing
[J].
Do the intestinal microbiotas differ between paddlefish (Polyodon spathala) and bighead carp (Aristichthys nobilis) reared in the same pond?
[J].A study was conducted to compare the intestinal microbial compositions of two fish species with similar feeding strategy; paddlefish (Polyodon spathala) and bighead carp (Aristichthys nobilis) reared in the same pond.Age-0 paddlefish and bighead carp with mean average body lengths of 43·39 ± 2·78 and 19·33 ± 3·68 cm, respectively, were reared with natural prey items in the same pond (20 m(2)). After 30 days of rearing, the intestinal microbiota of the two fish species was assessed by pyrosequencing of 16S rRNA genes. Interestingly, deviations were observed in the microbial communities of the two fish species according to the alpha- and beta-diversity measurements and detrended correspondence analysis (DCA). Shannon diversity (P = 0·015) and Pielou.evenness (P = 0·035) revealed significant lower diversity of the intestinal microbiota of paddlefish. Moreover, different core intestinal microbiota was noticed in the two fish species. Proteobacteria (57·3%), Firmicutes (11·9%), Fusobacteria (8·9%), Planctomycetes (7·3%), Actinobacteria (6·0%) and Verrucomicrobia (3·2%) were detected in bighead carp, while the dominant phyla in paddlefish intestines were Bacteroidetes (37·0%), Fusobacteria (35·1%), Firmicutes (14·8%) and Proteobacteria (12·6%).Our results revealed that the intestinal microbiota differed between paddlefish and bighead carp reared in the same pond when fed similar nature food. The potential host factors, such as the genetic background, gut histology and physiology are assumed to be involved in the intestinal bacterial compositions.Considering the similar feeding strategy of paddlefish and bighead carp, this study presents basic knowledge for evaluation of the importance of host factors (genetic background and gut anatomy) on intestinal microbial composition.© 2014 The Society for Applied Microbiology.
A comparative study on microbiota from the intestine of Prussian carp (Carassius gibelio) and their aquatic environmental compartments, using different molecular methods
[J].The aim of this study was to evaluate, via various molecular methods, the possible correlations between microbial community structure of Prussian carp and the environmental compartments of their habitat.Microbial communities in the intestine and environmental compartments were studied using PCR-screening, cloning and next-generation high-throughput sequencing of the 16S ribosomal RNA genes. The 16S rDNA metagenomic sequencing showed higher bacterial diversity in comparison with clone libraries, while group-specific PCR showed positive detection of nine bacteria phyla. Proteobacteria, Bacteroidetes, Firmicutes, Cyanobacteria and Actinobacteria were most abundant both in the intestine and habitat environments. The comparative analyses reveal that the bacterial community in the Prussian carp intestine is most similar to that identified from the chironomid.This study demonstrated some differences between molecular methods and showed advantages and limitations associated with them. These differences have the potential to reduce bias in results obtained from analysis of the community structure. The advantages of each molecular technique can be used for a better understanding of microbial diversity. The microbiota of Prussian carp intestine is most similar to those from the chironomids.We investigated the diversity of the intestinal microbiota in an economically important aquaculture species, the Prussian carp (Carassius gibelio). The results provide significant information to discuss possible functions of these bacteria for further understanding of Prussian carp health.© 2015 The Society for Applied Microbiology.
Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages
[J].The gut microbiota plays a significant role in the progression of fatty liver disease; however, the mediators and their mechanisms remain to be elucidated. Comparing metabolite profile differences between germ-free and conventionally raised mice against differences between mice fed a low- and high-fat diet (HFD), we identified tryptamine and indole-3-acetate (I3A) as metabolites that depend on the microbiota and are depleted under a HFD. Both metabolites reduced fatty-acid- and LPS-stimulated production of pro-inflammatory cytokines in macrophages and inhibited the migration of cells toward a chemokine, with I3A exhibiting greater potency. In hepatocytes, I3A attenuated inflammatory responses under lipid loading and reduced the expression of fatty acid synthase and sterol regulatory element-binding protein-1c. These effects were abrogated in the presence of an aryl-hydrocarbon receptor (AhR) antagonist, indicating that the effects are AhR dependent. Our results suggest that gut microbiota could influence inflammatory responses in the liver through metabolites engaging host receptors.Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Extensive impact of non-antibiotic drugs on human gut bacteria
[J].
Bacterial population in intestines of Litopenaeus vannamei fed different probiotics or probiotic supernatant
[J].
Effects of lactic acid bacteria and the corresponding supernatant on the survival, growth performance, immune response and disease resistance of Litopenaeus vannamei
[J].
Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota
[J].Animals have developed the means for supporting complex and dynamic consortia of microorganisms during their life cycle. A transcendent view of vertebrate biology therefore requires an understanding of the contributions of these indigenous microbial communities to host development and adult physiology. These contributions are most obvious in the gut, where studies of gnotobiotic mice have disclosed that the microbiota affects a wide range of biological processes, including nutrient processing and absorption, development of the mucosal immune system, angiogenesis, and epithelial renewal. The zebrafish (Danio rerio) provides an opportunity to investigate the molecular mechanisms underlying these interactions through genetic and chemical screens that take advantage of its transparency during larval and juvenile stages. Therefore, we developed methods for producing and rearing germ-free zebrafish through late juvenile stages. DNA microarray comparisons of gene expression in the digestive tracts of 6 days post fertilization germ-free, conventionalized, and conventionally raised zebrafish revealed 212 genes regulated by the microbiota, and 59 responses that are conserved in the mouse intestine, including those involved in stimulation of epithelial proliferation, promotion of nutrient metabolism, and innate immune responses. The microbial ecology of the digestive tracts of conventionally raised and conventionalized zebrafish was characterized by sequencing libraries of bacterial 16S rDNA amplicons. Colonization of germ-free zebrafish with individual members of its microbiota revealed the bacterial species specificity of selected host responses. Together, these studies establish gnotobiotic zebrafish as a useful model for dissecting the molecular foundations of host-microbial interactions in the vertebrate digestive tract.
The presence or absence of intestinal microbiota affects lipid deposition and related genes expression in zebrafish (Danio rerio)
[J].Understanding how intestinal microbiota alters energy homeostasis and lipid metabolism is a critical process in energy balance and health. However, the exact role of intestinal microbiota in the regulation of lipid metabolism in fish remains unclear. Here, we used two zebrafish models (germ-free and antibiotics-treated zebrafish) to identify the role of intestinal microbiota in lipid metabolism. Conventional and germ-free zebrafish larvae were fed with egg yolk. Transmission electron microscopy was used to detect the presence of lipid droplets in the intestinal epithelium. The results showed that, microbiota increased lipid accumulation in the intestinal epithelium. The mRNA sequencing technology was used to assess genes expression level. We found majority of the differentially expressed genes were related to lipid metabolism. Due to the limitation of germ-free zebrafish larvae, antibiotics-treated zebrafish were also used to identify the relationship between the gut microbiota and the host lipid metabolism. Oil-red staining showed antibiotics-treated zebrafish had less intestinal lipid accumulation than control group. The mRNA expression of genes related to lipid metabolism in liver and intestine was also quantified by using real-time PCR. The results indicated that apoa4, hsl, cox15, slc2a1a, and lss were more related to intestinal bacteria in fish, while the influence of intestinal microbiota on the activity of fabp6, acsl5, cd36, and gpat2 was different between the liver and intestine. This study identified several genes regulated by intestinal microbiota. Furthermore, the advantages and disadvantages of each model have been discussed. This study provides valuable information for exploring host-microbiota interactions in zebrafish in future.
Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection
[J].The gut microbiotas of zebrafish and mice share six bacterial divisions, although the specific bacteria within these divisions differ. To test how factors specific to host gut habitat shape microbial community structure, we performed reciprocal transplantations of these microbiotas into germ-free zebrafish and mouse recipients. The results reveal that communities are assembled in predictable ways. The transplanted community resembles its community of origin in terms of the lineages present, but the relative abundance of the lineages changes to resemble the normal gut microbial community composition of the recipient host. Thus, differences in community structure between zebrafish and mice arise in part from distinct selective pressures imposed within the gut habitat of each host. Nonetheless, vertebrate responses to microbial colonization of the gut are ancient: Functional genomic studies disclosed shared host responses to their compositionally distinct microbial communities and distinct microbial species that elicit conserved responses.
Gut microbiota in the pathogenesis of inflammatory bowel disease
[J].Inflammatory bowel disease (IBD), including ulcerative colitis and Crohn's disease, is a chronic and relapsing inflammatory disorder of the intestine. Although its incidence is increasing globally, the precise etiology remains unclear and a cure for IBD has yet to be discovered. The most accepted hypothesis of IBD pathogenesis is that complex interactions between genetics, environmental factors, and the host immune system lead to aberrant immune responses and chronic intestinal inflammation. The human gut harbors a complex and abundant aggregation of microbes, collectively referred to as the gut microbiota. The gut microbiota has physiological functions associated with nutrition, the immune system, and defense of the host. Recent advances in next-generation sequencing technology have identified alteration of the composition and function of the gut microbiota, which is referred to as dysbiosis, in IBD. Clinical and experimental data suggest dysbiosis may play a pivotal role in the pathogenesis of IBD. This review is focused on the physiological function of the gut microbiota and the association between the gut microbiota and pathogenesis in IBD. In addition, we review the therapeutic options for manipulating the altered gut microbiota, such as probiotics and fecal microbiota transplantation.
Role of gastrointestinal microbiota in fish
[J].
Enzyme production by obligate intestinal anaerobic bacteria isolated from oscars(Astronotus ocellatus), angelfish (Pterophyllum scalare) and southern flounder (Paralichthys lethostigma)
[J].
Antibacterial abilities of intestinal bacteria in freshwater cultured fish
[J].
The evolution of the host microbiome as an ecosystem on a leash
[J].
Innate immune signaling in defense against intestinal microbes
[J].The gastrointestinal system is a common entry point for pathogenic microbes to access the inner environment of the body. Anti-microbial factors produced by the intestinal mucosa limit the translocation of both commensal and pathogenic microbes across the intestinal epithelial cell barrier. The regulation of these host defense mechanisms largely depends on the activation of innate immune receptors by microbial molecules. Under steady-state conditions, the microbiota provides constitutive signals to the innate immune system, which helps to maintain a healthy inflammatory tone within the intestinal mucosa and, thus, enhances resistance to infection with enteric pathogens. During an acute infection, the intestinal epithelial cell barrier is breached, and the detection of microbial molecules in the intestinal lamina propria rapidly stimulates innate immune signaling pathways that coordinate early defense mechanisms. Herein, we review how microbial molecules shed by both commensal and pathogenic microbes direct host defenses at the intestinal mucosa. We highlight the signaling pathways, effector molecules, and cell populations that are activated by microbial molecule recognition and, thereby, are involved in the maintenance of homeostatic levels of host defense and in the early response to acute enteric infection. Finally, we discuss how manipulation of these host defense pathways by stimulating innate immune receptors is a potential therapeutic strategy to prevent or alleviate intestinal disease.© 2011 John Wiley & Sons A/S.
Research advances on probiotics and fish gut health
[J].
Cupriavidus in the intestinal microbiota of Tibet endemic fish Glyptosternum maculatum can help it adapt to habitat of the Qinghai Tibet Plateau
[J].Despite extensive research, many questions remain unanswered about common problems that impact dog welfare, particularly where there are multiple contributing factors that can occur months or years before the problem becomes apparent. The Generation Pup study is the first longitudinal study of dogs that recruits pure- and mixed-breed puppies, aiming to investigate the relative influence of environmental and genetic factors on a range of health and behaviour outcomes, (including separation related behaviour, aggression to familiar/unfamiliar people or dogs and obesity). This paper describes the study protocol in detail.
Characterization of the skin and gill microbiomes of the farmed seabass (Dicentrarchus labrax) and seabream (Sparus aurata)
[J].